Recently I have a friend asking me how to fit a function to some observational data using python. Well, it depends on whether you have a function form in mind. If you have one, then it is easy to do that. But even you don’t know the form of the function you want to fit, you can still do it fairly easy. Here are some examples. You can find all the code on Qingkai’s Github.
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
plt.style.use('seaborn-poster')
If you can tell the function form from the data
For example, if I have the following data that actually generated from the function 3*exp(-0.05x) + 12, but with some noise. When we see this dataset, we can tell it might be generated from an exponential function.
np.random.seed(42)
x = np.arange(-10, 10, 0.1)
# true data generated by this function
y = 3 * np.exp(-0.05*x) + 12
# adding noise to the true data
y_noise = y + np.random.normal(0, 0.2, size = len(y))
plt.figure(figsize = (10, 8))
plt.plot(x, y_noise, 'o', label = 'Raw data generated')
plt.xlabel('X')
plt.ylabel('y')
plt.legend()
plt.show()
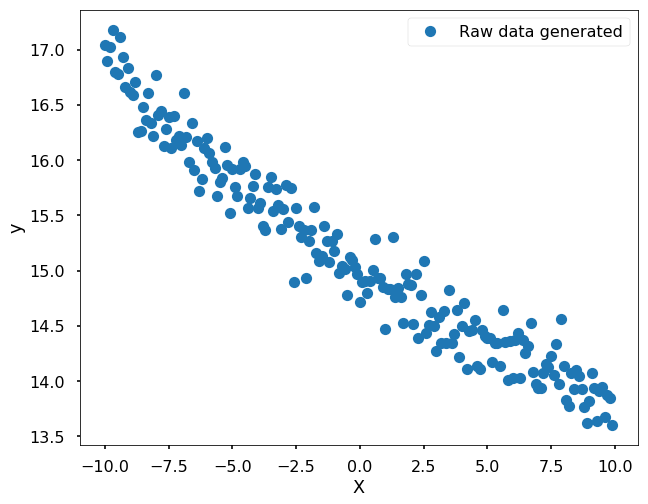
Since we have the function form in mind already, let’s fit the data using scipy function - curve_fit
from scipy.optimize import curve_fit
def func(x, a, b, c):
return a * np.exp(-b * x) + c
popt, pcov = curve_fit(func, x, y_noise)
plt.figure(figsize = (10, 8))
plt.plot(x, y_noise, 'o',
label='Raw data')
plt.plot(x, func(x, *popt), 'r',
label='Fit: a=%5.3f, b=%5.3f, c=%5.3f' % tuple(popt))
plt.legend()
plt.xlabel('X')
plt.ylabel('y')
plt.show()
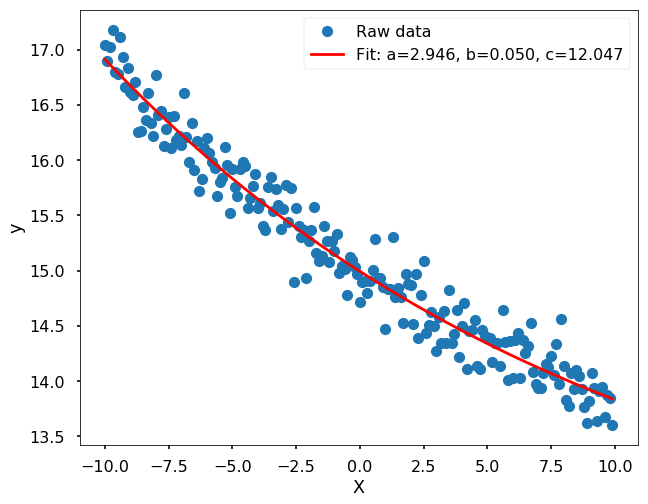
More complicated case, you don’t know the funciton form
For a more complicated case that we can not easily guess the form of the function, we could use a Spline to fit the data. For example, we could use UnivariateSpline.
from scipy.interpolate import UnivariateSpline
np.random.seed(42)
x = np.arange(-10, 10, 0.1)
# true data generated by this function
y = 3 * np.exp(-0.05*x) + 12 + 1.4 * np.sin(1.2*x) + 2.1 * np.sin(-2.2*x + 3)
# adding noise to the true data
y_noise = y + np.random.normal(0, 0.5, size = len(y))
plt.figure(figsize = (10, 8))
plt.plot(x, y_noise, 'o', label = 'Raw data generated')
plt.plot(x, y, 'k', label = 'True model')
plt.xlabel('X')
plt.ylabel('y')
plt.legend()
plt.show()
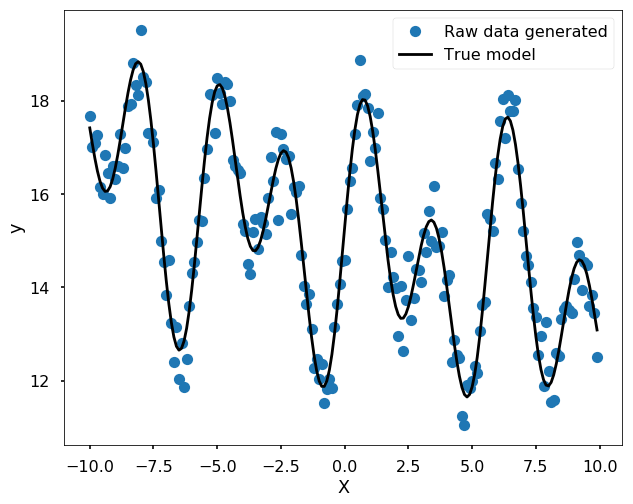
# Note, you need play with the s - smoothing factor
s = UnivariateSpline(x, y_noise, s=15)
xs = np.linspace(-10, 10, 100)
ys = s(xs)
plt.figure(figsize = (10, 8))
plt.plot(x, y_noise, 'o', label = 'Raw data')
plt.plot(x, y, 'k', label = 'True model')
plt.plot(xs, ys, 'r', label = 'Fitting')
plt.xlabel('X')
plt.ylabel('y')
plt.legend(loc = 1)
plt.show()
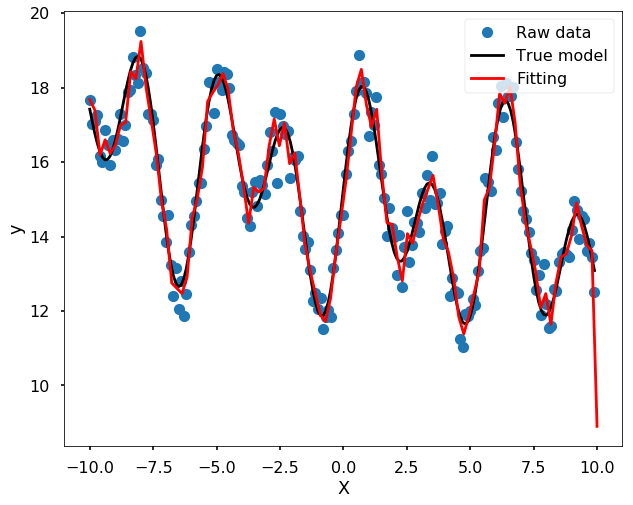
Of course, you could also use Machine Learning algorithms
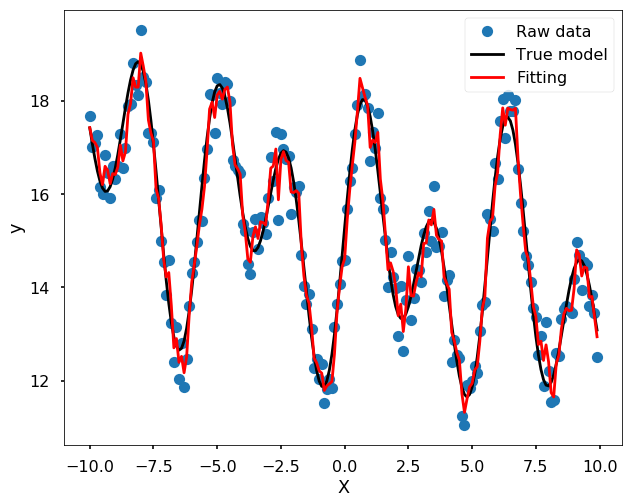
Many machine learning algorithms could do the job as well, you could treat this as a regression problem in machine learning, and train some model to fit the data well. I will show you two methods here - Random forest and ANN. I didn’t do any test here, it is only fitting the data and use my sense to choose the parameters to avoid the model too flexible.
Use Random Forest
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error
# fit the model and get the estimation for each data points
yfit = RandomForestRegressor(50, random_state=42).fit(x[:, None], y_noise).predict(x[:, None])
plt.figure(figsize = (10,8))
plt.plot(x, y_noise, 'o', label = 'Raw data')
plt.plot(x, y, 'k', label = 'True model')
plt.plot(x, yfit, '-r', label = 'Fitting', zorder = 10)
plt.legend()
plt.xlabel('X')
plt.ylabel('y')
plt.show()
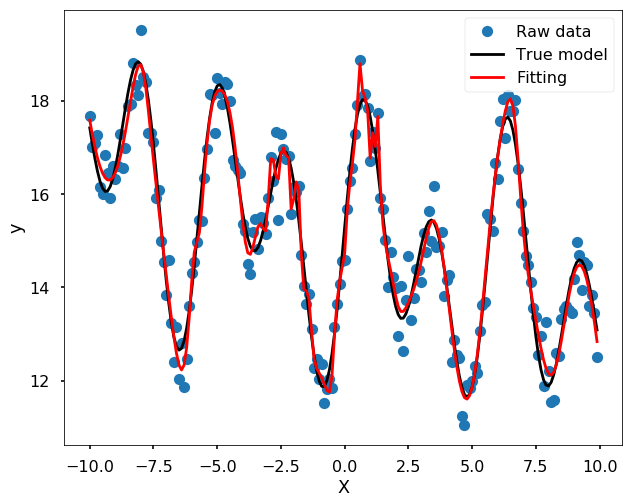
Use ANN
from sklearn.neural_network import MLPRegressor
mlp = MLPRegressor(hidden_layer_sizes=(100,100,100), max_iter = 5000, solver='lbfgs', alpha=0.01, activation = 'tanh', random_state = 8)
yfit = mlp.fit(x[:, None], y_noise).predict(x[:, None])
plt.figure(figsize = (10,8))
plt.plot(x, y_noise, 'o', label = 'Raw data')
plt.plot(x, y, 'k', label = 'True model')
plt.plot(x, yfit, '-r', label = 'Fitting', zorder = 10)
plt.legend()
plt.xlabel('X')
plt.ylabel('y')
plt.show()
I discovered your this post while scanning for some related data on online journal search...Its a decent post..keep posting and upgrade the datafriv Games online 2020
ReplyDeleteJogos live
friv game
Curve fitting is the process of constructing a mathematical function that best fits a given set of data points. In Python, libraries like NumPy and SciPy offer robust tools for this task.
DeleteMachine Learning Final Year Projects
Key Concepts
Data Points: The observed values (x, y) that need to be approximated by a function.
Model Function: The mathematical function used to fit the data. Examples include linear, polynomial, exponential, and logarithmic functions.
Optimization: The process of finding the parameters of the model function that minimize the difference between the fitted curve and the data points.
python projects for final year students
Goodness of Fit: Measures how well the fitted curve represents the data. Common metrics include R-squared, Mean Squared Error (MSE), and Root Mean Squared Error (RMSE).
Python Libraries
NumPy: Provides essential numerical operations and array manipulation.
SciPy: Offers optimization functions, including curve_fit for nonlinear least squares fitting.
Matplotlib: For visualizing the data and the fitted curve.
Deep Learning Projects for Final Year Students
Amazing - with your random forest example I finally got my dataset to predict y values. Thankyou for writing this!
ReplyDeleteInternet Business Ideas for 2020
ReplyDeleteAm here to testify what this great spell caster done for me. i never believe in spell casting, until when i was was tempted to try it. i and my wife have been having a lot of problem living together, she will always not make me happy because she have fallen in love with another man outside our relationship, i tried my best to make sure that my wife leave this woman but the more i talk to her the more she makes me fell sad, so my marriage is now leading to divorce because she no longer gives me attention. so with all this pain and agony, i decided to contact this spell caster to see if things can work out between me and my wife again. this spell caster who was a man told me that my wife is really under a great spell that she have been charm by some magic, so he told me that he was going to make all things normal back. he did the spell on my wife and after 5 days my wife changed completely she even apologize with the way she treated me that she was not her self, i really thank this man his name is Dr ose he have bring back my wife back to me i want you all to contact him who are having any problem related to marriage issue and relationship problem he will solve it for you. his email is oseremenspelltemple@gmail.com he is a man and his great. wish you good time.
ReplyDeleteHe cast spells for different purposes like
(1) If you want your ex back.
(2) if you always have bad dream
(3) You want to be promoted in your office.
(4) You want women/men to run after you.
(5) If you want a child.
(6) You want to be rich.
(7) You want to tie your husband/wife to be yours forever.
(8) If you need financial assistance.
(9) HIV/AIDS CURE
(10) is the only answer to that your problem of winning the lottery
Contact him today on oseremenspelltemple@gmail.com or whatsapp him on +2348136482342
Five weeks ago my boyfriend broke up with me. It all started when i went to summer camp i was trying to contact him but it was not going through. So when I came back from camp I saw him with a young lady kissing in his bed room, I was frustrated and it gave me a sleepless night. I thought he will come back to apologies but he didn't come for almost three week i was really hurt but i thank Dr.Azuka for all he did i met Dr.Azuka during my search at the internet i decided to contact him on his email dr.azukasolutionhome@gmail.com he brought my boyfriend back to me just within 48 hours i am really happy. What’s app contact : +44 7520 636249
ReplyDeleteMy name is Tom cam!!! i am very grateful sharing this great testimonies with you all, The best thing that has ever happened in my life, is how i worn the Powerball lottery. I do believe that someday i will win the Powerball lottery. finally my dreams came through when i contacted Dr. OSE and tell him i needed the lottery winning special numbers cause i have come a long way spending money on ticket just to make sure i win. But i never knew that winning was so easy with the help of Dr. OSE, until the day i meant the spell caster testimony online, which a lot of people has talked about that he is very powerful and has great powers in casting lottery spell, so i decided to give it a try. I emailed Dr. OSE and he did a spell and gave me the winning lottery special numbers 62, and co-incidentally I have be playing this same number for the past 23years without any winning, But believe me when I play the special number 62 this time and the draws were out i was the mega winner because the special 62 matched all five white-ball numbers as well as the Powerball, in the April 4 drawing to win the $70 million jackpot prize...… Dr. OSE, truly you are the best, with Dr. OSE you can will millions of money through lottery. i am a living testimony and so very happy i meant him, and i will forever be grateful to him...… you can Email him for your own winning special lottery numbers now oseremenspelltemple@gmail.com OR WHATSAPP him +2348136482342
ReplyDeleteDR EMU WHO HELP PEOPLE IN ANY TYPE OF LOTTERY NUMBERS
ReplyDeleteIt is a very hard situation when playing the lottery and never won, or keep winning low fund not up to 100 bucks, i have been a victim of such a tough life, the biggest fund i have ever won was 100 bucks, and i have been playing lottery for almost 12 years now, things suddenly change the moment i came across a secret online, a testimony of a spell caster called DR EMU, who help people in any type of lottery numbers, i was not easily convinced, but i decided to give try, now i am a proud lottery winner with the help of DR EMU, i won $1,000.0000.00 and i am making this known to every one out there who have been trying all day to win the lottery, believe me this is the only way to win the lottery.
Contact him via email Emutemple@gmail.com
What's app +2347012841542
Https://emutemple.wordpress.com/
I am so stoked to have found your weblog, I stumbled here by accident while researching for something else, Regardless I am here now and would just like to say thank you for this fantastic post and all round entertaining blog (I also love the theme/design), We have come up with ways on HOW TO BUY DATABASE CERTIFICATES ONLINE
ReplyDeleteI don't have time to go through it all at the minute but I have bookmarked it and also added your RSS feeds, so when I have time I will be back to read a lot more, Please do keep up the excellent job.
Are you in need of Loan? Here all problem regarding Loans is solve between a short period of time what are you waiting for apply now and solve your problem or start a business with funds Contact us now. many more 2% interest rate.(Whats App) number +919394133968 patialalegitimate515@gmail.com
ReplyDeleteMr Sorina