We talked about the basics of SVM in the previous blog, and now let's use an example to show you how to use it easily with sklearn, and also, some of the important parameters of it. You can find all the code at Qingkai's Github.
import numpy as np
import itertools
import matplotlib.pyplot as plt
from sklearn import svm, datasets
from sklearn.cross_validation import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import classification_report,confusion_matrix
plt.style.use('seaborn-poster')
%matplotlib inlineLoad and visualize data
Let's load the IRIS from sklearn. This dataset is a very famous dataset for pattern recognition that from the famous Fisher's paper in 1936. The data set contains 3 classes of 50 instances each, where each class refers to a type of iris plant. The features are Sepal Length, Sepal Width, Petal Length and Petal Width. They are related with the physical properties of the plant. Today we will use only 2 features for easy visualization purposes.
# import the iris data
iris = datasets.load_iris()
# let's just use the first two features, so that we can
# easily visualize them
X = iris.data[:, [0, 2]]
y = iris.target
target_names = iris.target_names
feature_names = iris.feature_names
# get the classes
n_class = len(set(y))
print('We have %d classes in the data'%(n_class))We have 3 classes in the data# let's have a look of the data first
plt.figure(figsize = (10,8))
for i, c, s in (zip(range(n_class), ['b', 'g', 'r'], ['o', '^', '*'])):
ix = y == i
plt.scatter(X[:, 0][ix], X[:, 1][ix], color = c, marker = s, s = 60, label = target_names[i])
plt.legend(loc = 2, scatterpoints = 1)
plt.xlabel('Feature 1 - ' + feature_names[0])
plt.ylabel('Feature 2 - ' + feature_names[2])
plt.show()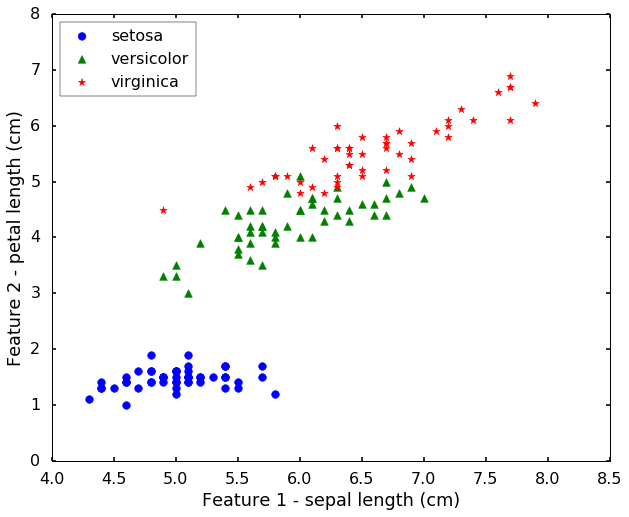
Train an SVM classifier
Since our purpose is to understand the SVM classifier, therefore, we won't split the data into training, validation, and test dataset. Also, we will not scale the data, because the values are all similar. (In reality, you still want this in your workflow, and use cross-validation or gridsearch to determine some parameters). Here, let's just train the SVM classifier directly.
# Initialize SVM classifier
clf = svm.SVC(kernel='linear')
# Train the classifier with data
clf.fit(X,y)SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape=None, degree=3, gamma='auto', kernel='linear',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False)The training results
Let's view the performance on the training data, we will plot the confusion matrix. Also, we will plot the decision boundary, which will help us understand more of the capability of the classifier (since we only have two selected features, this makes us easier to view the decision boundary).
def plot_confusion_matrix(cm, classes,
normalize=False,
title='Confusion matrix',
cmap=plt.cm.Blues):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
fig = plt.figure(figsize=(10, 8))
plt.imshow(cm, interpolation='nearest', cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
if normalize:
cm = cm.astype('float') / cm.sum(axis=1)[:, np.newaxis]
thresh = cm.max() / 2.
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(j, i, cm[i, j],
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black")
plt.tight_layout()
plt.ylabel('True label')
plt.xlabel('Predicted label')
plt.show()
# Plotting decision regions
def plot_desicion_boundary(X, y, clf, title = None):
'''
Helper function to plot the decision boundary for the SVM
'''
x_min, x_max = X[:, 0].min() - 1, X[:, 0].max() + 1
y_min, y_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.1),
np.arange(y_min, y_max, 0.1))
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.figure(figsize = (10, 8))
plt.contourf(xx, yy, Z, alpha=0.4)
plt.scatter(X[:, 0], X[:, 1], c=y, alpha=0.8)
if title is not None:
plt.title(title)
# highlight the support vectors
#plt.scatter(clf.support_vectors_[:, 0], clf.support_vectors_[:, 1], s=80,
# facecolors='none', zorder=10)
plt.xlabel('Feature 1')
plt.ylabel('Feature 2')
plt.show()# predict results from the test data
predicted = clf.predict(X)
# plot the confusion matrix
cm = confusion_matrix(y,predicted)
plot_confusion_matrix(cm, classes=iris.target_names,
title='Confusion matrix, without normalization')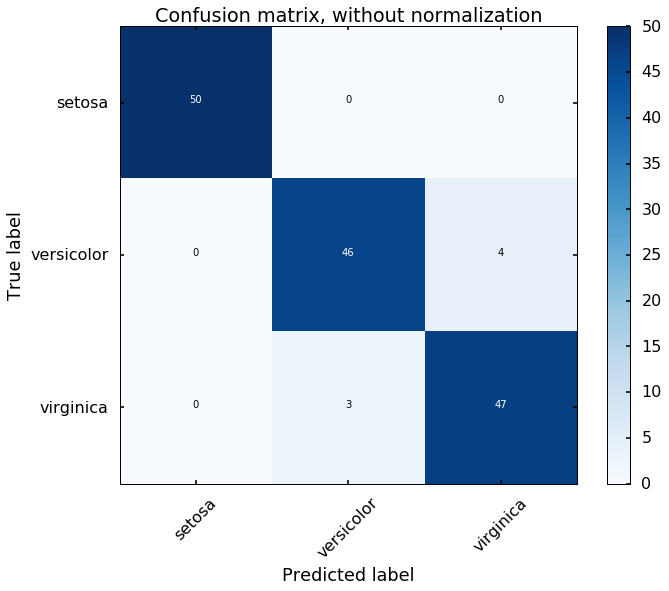
plot_desicion_boundary(X, y, clf)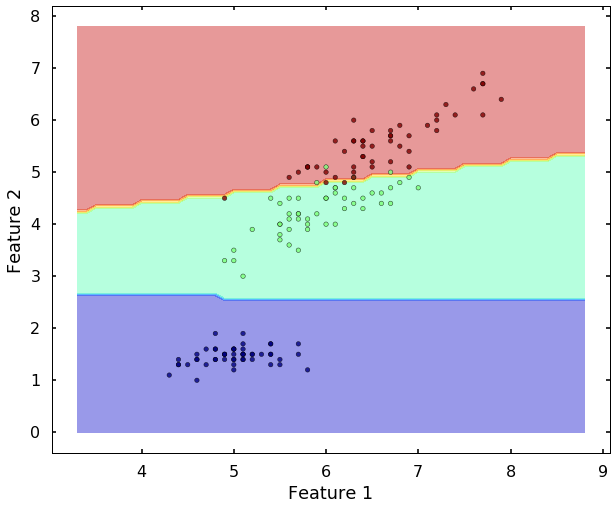
We can see the above two figures, the SVM classifier actually did a decent job to separate different classes. Since the data is linearly separatable, the 'linear' kernel can satisfy the job. But what if the data is not linearly separatable? Let's see another example.
Non-linearly separatable example
# let's construct a dataset that not linearly separatable
X1, y1 = datasets.make_gaussian_quantiles(cov=2.,
n_samples=400, n_features=2,
n_classes=2, random_state=1)
X2, y2 = datasets.make_gaussian_quantiles(mean=(3, 3), cov=1.5,
n_samples=400, n_features=2,
n_classes=2, random_state=1)
X = np.concatenate((X1, X2))
y = np.concatenate((y1, - y2 + 1))
n_class = 2# let's have a look of the data first
plt.figure(figsize = (10,8))
for i, c, s in (zip(range(n_class), ['b', 'g'], ['o', '^'])):
ix = y == i
plt.scatter(X[:, 0][ix], X[:, 1][ix], color = c, marker = s, s = 60)
plt.xlabel('Feature 1')
plt.ylabel('Feature 2')
plt.show()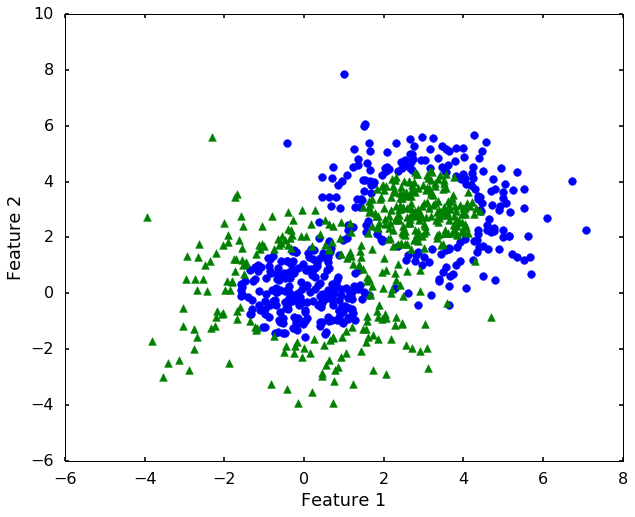
clf = svm.SVC(kernel='linear')
clf.fit(X,y)SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape=None, degree=3, gamma='auto', kernel='linear',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False)plot_desicion_boundary(X, y, clf)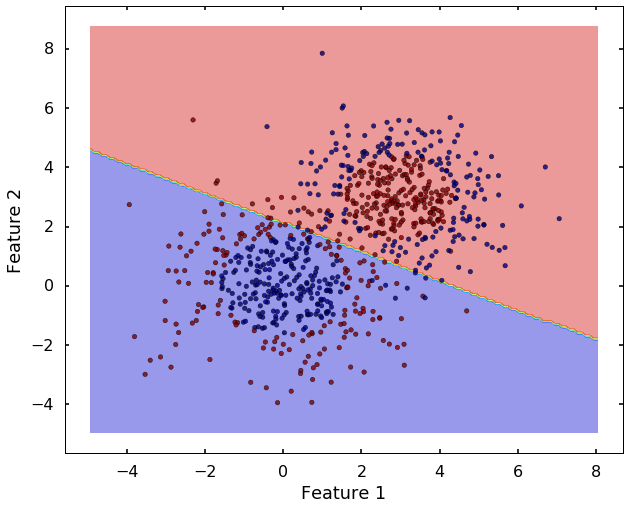
Now we can see that, the 'linear' kernel is not working anymore. It tries to use a linear boundary to classify the data, which is impossible. Remembered that we talked in the previous blog that using a different kernel will transform this data into higher dimensions to separate them linearly. There are a couple of options in sklearn to choose, the most popular ones are 'rbf' - radial basis function, 'poly' - the polynomial. We will use 'rbf' here.
clf = svm.SVC(kernel='rbf')
clf.fit(X,y)SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape=None, degree=3, gamma='auto', kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False)plot_desicion_boundary(X, y, clf)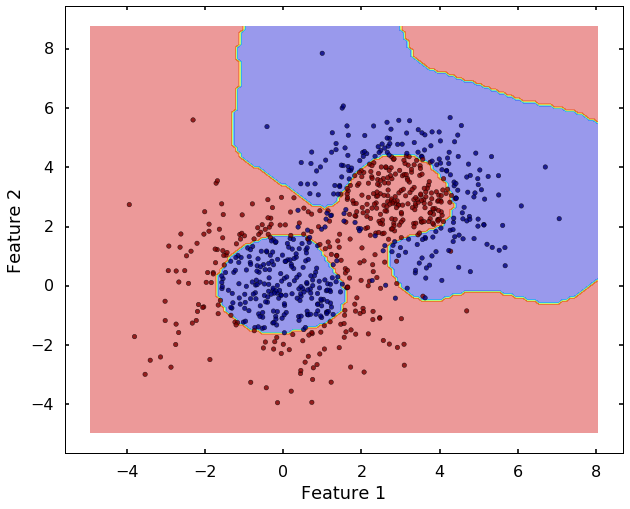
Haha, after we used the 'rbf' kernel, it seems the SVM can classify them much better. The 'rbf' kernel based SVM looks much flexible, but it also more prone to overfit the data. Therefore, to better use it, you need understand two important parameters: C and gamma, let's have a closer look at them.
C and gamma in SVM
The C and gamma parameter in the SVM are the most important parameters for the 'rbf' kernel, that can be determined by the grid-search method. But we need get a sense how they affect the results if we change them.
C controls the cost of misclassification on the training data, it trades off misclassification on training examples against simplicity of the decision boundary. A large C makes the cost of misclassification high, this will force the algorithm to fit the data with more flexible model, and try to classify the training data correctly as much as possible. But this will also make the SVM more easier to overit the data. Therefore, the simple rule to remember is:
- Larger C, lower bias, higher variance.
- Smaller C, higher bias, lower variance.
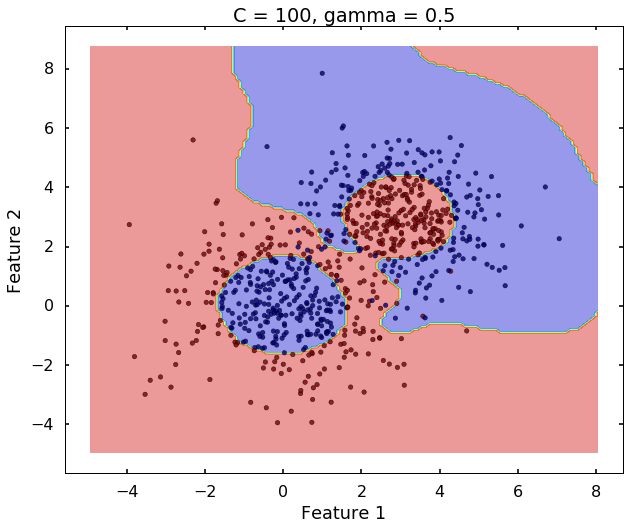
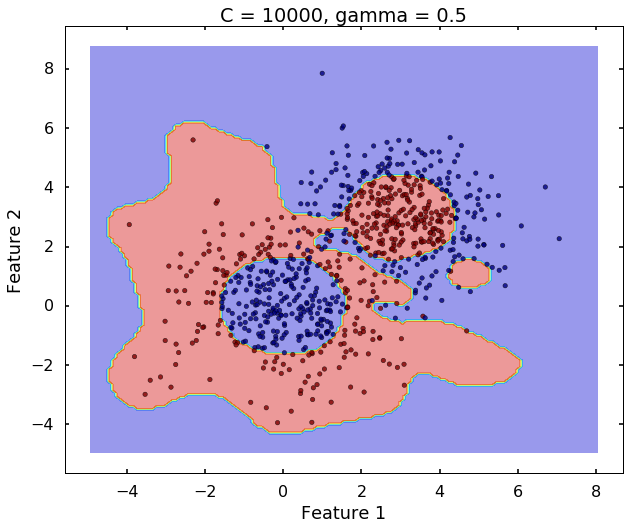
The following 3 figures showing different values of C, that is, C = 1, C = 100, C = 10000. We can see the higher the value C, the more complicated the decision boundary is, which tries to fit every training data, even with some clearly anomaly data.
# C = 1
clf = svm.SVC(kernel='rbf', C = 1, gamma = 0.5)
clf.fit(X,y)
plot_desicion_boundary(X, y, clf, title = 'C = 1, gamma = 0.5')
# C = 100
clf = svm.SVC(kernel='rbf', C = 100, gamma = 0.5)
clf.fit(X,y)
plot_desicion_boundary(X, y, clf, title = 'C = 100, gamma = 0.5')
# C = 10000
clf = svm.SVC(kernel='rbf', C = 10000, gamma = 0.5)
clf.fit(X,y)
plot_desicion_boundary(X, y, clf, title = 'C = 10000, gamma = 0.5')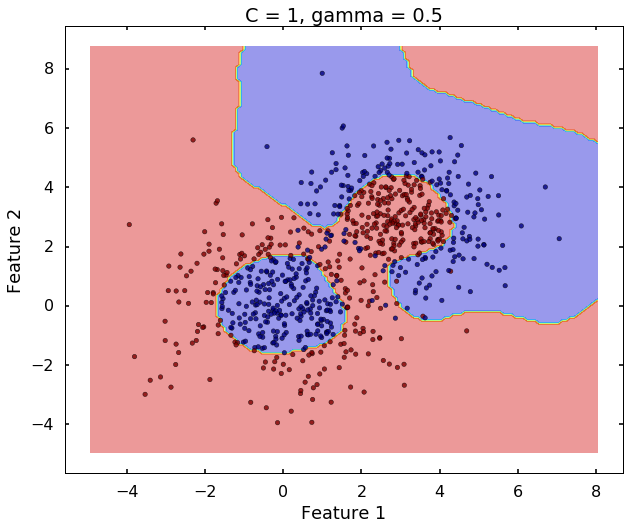
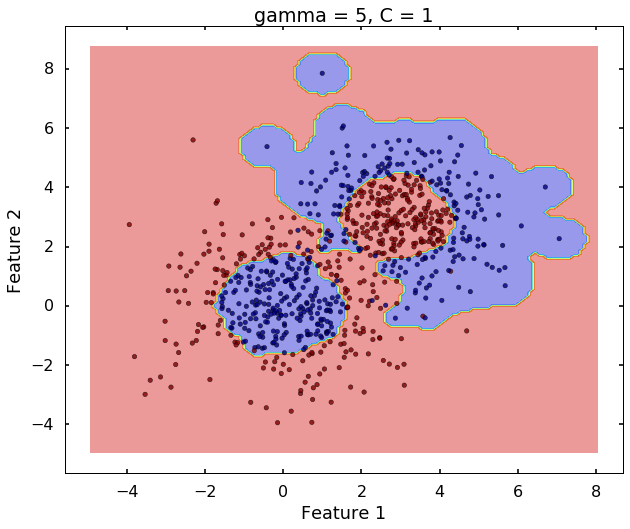
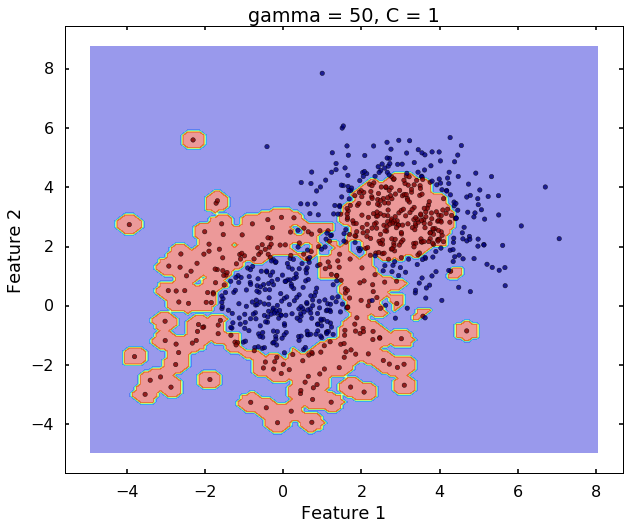
Technically, gamma is not a parameter of the SVM, but a parameter for the 'rbf' kernel to handle non-linear classification. It defines how far the influence of a single training example reaches. A low value of gamma means 'far' and high value means 'close'. Therefore, A small gamma means a radial basis function with far influence, i.e. if x is a support vector, a small gamma implies the class of this support vector will have influence on deciding the class of the vector y even if the distance between them is large. If gamma is large, then variance is small implying the support vector does not have wide-spread influence. A rule to remember:
- large gamma leads to high bias and low variance
- small gamma leads to low bias and high variance
# gamma = 0.5
clf = svm.SVC(kernel='rbf', gamma = 0.5, C = 1)
clf.fit(X,y)
plot_desicion_boundary(X, y, clf, title = 'gamma = 0.5, C = 1')
# gamma = 5
clf = svm.SVC(kernel='rbf', gamma = 5, C = 1)
clf.fit(X,y)
plot_desicion_boundary(X, y, clf, title = 'gamma = 5, C = 1')
# gamma = 50
clf = svm.SVC(kernel='rbf', gamma = 50, C = 1)
clf.fit(X,y)
plot_desicion_boundary(X, y, clf, title = 'gamma = 50, C = 1')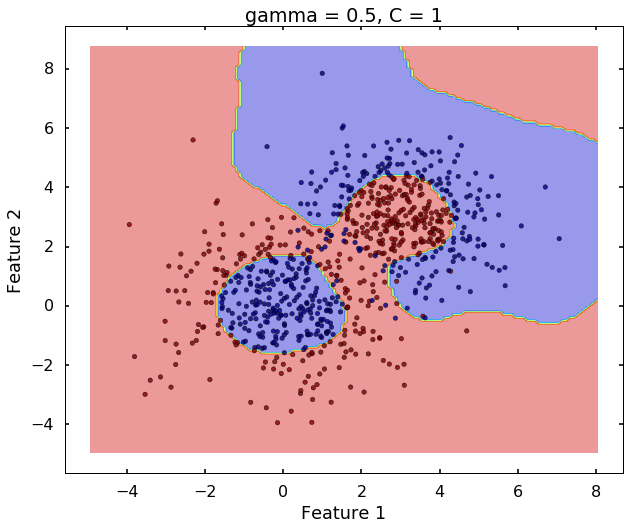
More on the parameters
Another very important parameter is the class_weight, which will address the inbalance class case. Let's see an extreme example: if we have 99 instances of class 0, but 1 instance of class 1, then even the algorithm classify everything to class 0, we will have 99% correct rate. This is always the case if we have an inbalanced dataset, there are many ways to deal with it, but the parameter 'class_weight' in SVM will deal with this case by setting different weights to different classes. Let's grab the example from sklearn website directly. We can see without specify the class_weight, the boundary is set where most of the blue dots on beneath the boundary. But after we specified the class_weight, the SVM now pays more attention to the minor class (class 1), and classify most of them correct.
# we create 1000 data points for class 0
# and 100 data points for class 1
rng = np.random.RandomState(0)
n_samples_1 = 1000
n_samples_2 = 100
X = np.r_[1.5 * rng.randn(n_samples_1, 2),
0.5 * rng.randn(n_samples_2, 2) + [2, 2]]
y = [0] * (n_samples_1) + [1] * (n_samples_2)
# fit the model and get the separating hyperplane
clf = svm.SVC(kernel='linear', C=1.0)
clf.fit(X, y)
w = clf.coef_[0]
a = -w[0] / w[1]
xx = np.linspace(-5, 5)
yy = a * xx - clf.intercept_[0] / w[1]
# get the separating hyperplane using weighted classes
# note we give a smaller weight to class 0
wclf = svm.SVC(kernel='linear', class_weight={1: 10})
wclf.fit(X, y)
ww = wclf.coef_[0]
wa = -ww[0] / ww[1]
wyy = wa * xx - wclf.intercept_[0] / ww[1]
# plot separating hyperplanes and samples
plt.figure(figsize = (10, 8))
h0 = plt.plot(xx, yy, 'k-', label='no weights')
h1 = plt.plot(xx, wyy, 'k--', label='with weights')
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.Paired)
plt.legend()
plt.xlabel('Feature 1')
plt.ylabel('Feature 2')
plt.axis('tight')
plt.show()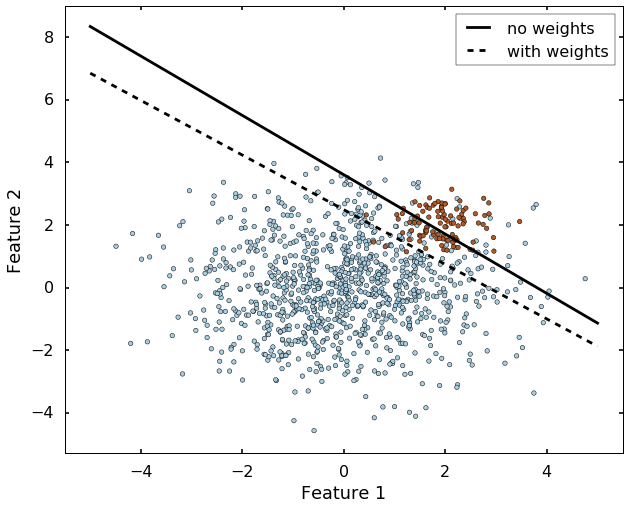
I have read your blog its very attractive and impressive. I like your blog.
ReplyDeletemachine learning online course
Intuitive Support Vector Machines (SVMs) refer to an approach or perspective that aims to simplify the understanding and application of Support Vector Machines, a powerful supervised learning algorithm used for classification and regression tasks.
DeleteMargin: SVMs aim to find the hyperplane that maximizes the margin between different classes of data points. The margin is the distance between the hyperplane and the nearest data points (support vectors). Maximizing this margin helps in achieving a good generalization capability of the model.
DeleteKernel Trick: SVMs can efficiently perform nonlinear classification using what's known as the kernel trick. This trick allows SVMs to implicitly map the input data into high-dimensional feature spaces without explicitly computing the transformations. This is particularly useful when the data is not linearly separable in its original feature space.
Machine Learning Final Year Projects
Took me time to understand all of the comments, but I seriously enjoyed the write-up. It proved being really helpful to me and Im positive to all of the commenters right here! Its constantly nice when you can not only be informed, but also entertained! I am certain you had enjoyable writing this write-up.
ReplyDelete360DigiTMG
Nice blog
ReplyDeleteangularjs online course
Angular Online Training
Awesome blog. I enjoyed reading your articles. This is truly a great read for me. I have bookmarked it and I am looking forward to reading new articles. Keep up the good work!
ReplyDeleteFive weeks ago my boyfriend broke up with me. It all started when i went to summer camp i was trying to contact him but it was not going through. So when I came back from camp I saw him with a young lady kissing in his bed room, I was frustrated and it gave me a sleepless night. I thought he will come back to apologies but he didn't come for almost three week i was really hurt but i thank Dr.Azuka for all he did i met Dr.Azuka during my search at the internet i decided to contact him on his email dr.azukasolutionhome@gmail.com he brought my boyfriend back to me just within 48 hours i am really happy. What’s app contact : +44 7520 636249
ReplyDeleteIm obliged for the blog article.Thanks Again. Awesome.
ReplyDeletemachine learning course in hyderabad
best machine learning course in india
Nice Content !If your business is having any issues you can dial the QuickBooks helpline on quickbooks customer service+1 855-428-7237
ReplyDelete